
As a conclusion, surveillance must be exercised on ruminant farms in the surroundings of the infected area in order to evaluate the possible extension of the disease and their potential role as reservoirs of T.
#Como calcular el intervalo de confianza free
According to FreeCalc assessment, cattle and goat populations would be free from disease however, the results from sheep are not adequate to conclude that the population would be free from disease. evansi could be demonstrated in none of them. Of a total of 1228 ruminants assessed, 61 (5%) were serologically positive (7 cattle, 21 goats, 33 sheep), but T. To evaluate possible reservoirs in the area a representative sample of domestic ruminants was examined by serological, parasitological and molecular tests. However, a little area remains still infected despite the use of the same control measures.

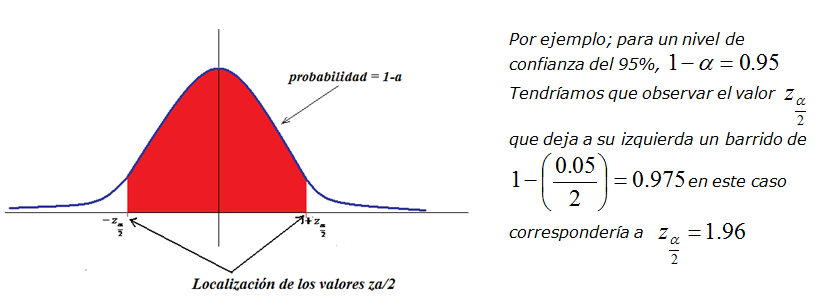
evansi from most of the infected areas in the Archipelago. Intervalo De Confianza, Quimica Analitica, Probabilidad Y Estadistica, Calculo, Frmulas Matemticas. The disease (surra) was first diagnosed in the Canary Islands in a dromedary camel in 1997 thus, a control plan was implemented achieving the eventual eradication of T. more Trypanosoma evansi is the most widely spread of the pathogenic African trypanosomes of animals. Trypanosoma evansi is the most widely spread of the pathogenic African trypanosomes of animals. En R existen diferentes métodos para poder realizarlo, aunque en esta entrada vamos a emplear el test binomial exacto con la función binom.test () del paquete stats. This article describes in detail how our package works for the desired applications, including illustrated explanations and easily reproducible examples. Calcular los intervalos de confianza para una proporción o un índice es muy sencillo. The alleHap package also has a function to simulate pedigrees including all these scenarios. Several scenarios are analyzed: family completely genotyped, children partially genotyped and parents completely genotyped, children fully genotyped and parents containing entirely or partially missing genotypes, and founders and their offspring both only partially genotyped. Existen tres factores que determinan la confianza de tu investigación: Tamaño de la muestra. La mayoría de los investigadores utilizan el nivel de confianza del 95. If haplotypes are identified in parents or offspring, missing alleles can be imputed in subjects containing missing values. El nivel de confianza se refiere a la tasa de éxito a largo plazo del método, es decir, la frecuencia con la que este tipo de intervalo capturará el parámetro de interés. When genotypes of related individuals are available in a number of linked genetic markers, the program starts by identifying haplotypes compatible with the observed genotypes in those markers without missing values. more The alleHap package is designed for imputing genetic missing data and reconstruct non-recombinant haplotypes from pedigree databases in a deterministic way.

The alleHap package is designed for imputing genetic missing data and reconstruct non-recombinant.


 0 kommentar(er)
0 kommentar(er)
